Paper is now live!

Go to Preprint: https://doi.org/10.1093/ismeco/ycaf094
The KSGP Database
Databases of Small Subunit (SSU) Ribosomal RNA sequences play a key role in the study of microbial communities, particularly metabarcoding of prokaryote communities. However, the RDP database is no longer being updated; the most recent releases of the Greenegenes and SILVA databases date from 2019, although Greengenes has recently be superseded by Greengenes2. Taxonomic annotations of environmental sequences in these databases are often incomplete and databases contain appreciable numbers of taxonomic errors (Edgar, 2018; Smith, Glendinning, Walker, & Watson, 2022).
Here we provide a quality controlled and deduplicated database of 16S sequences from the Genome Taxonomy Database (GTDB) version 214.0 (D. H. Parks, 2023; D. H. Parks et al., 2020; Donovan H Parks et al., 2021; D. H. Parks et al., 2018) from which sequences which appear to be assigned to the incorrect domain have been removed. We also provide the current version of our KSPG database, which uses these cleaned GTDB sequences combined with the PR2 database of eukaryote 18S sequences to annotate a large collection of near full length environmental rRNA sequences (Karst et al., 2018) and to re-annotated the Archaea sequences from SILVA version 138.1.
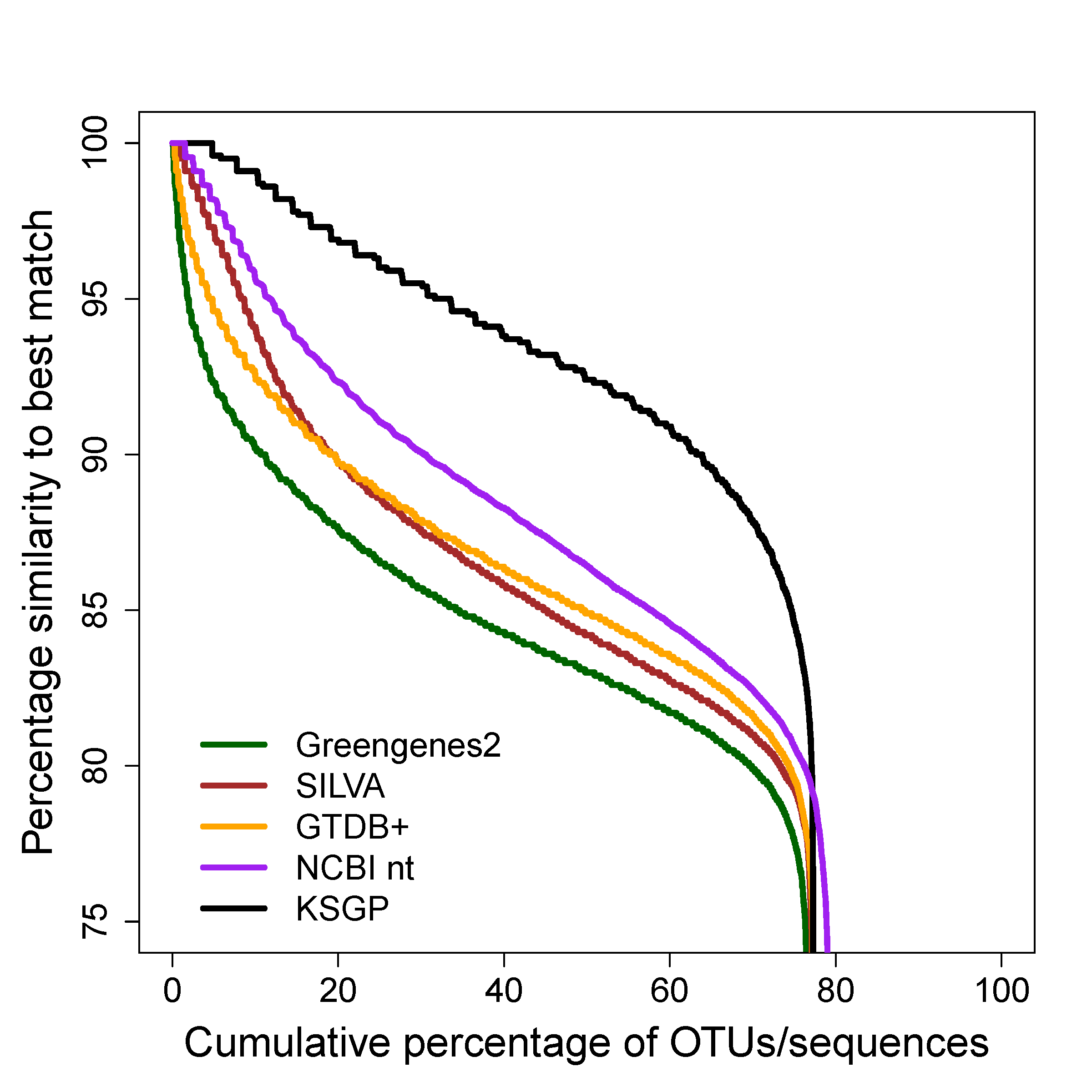
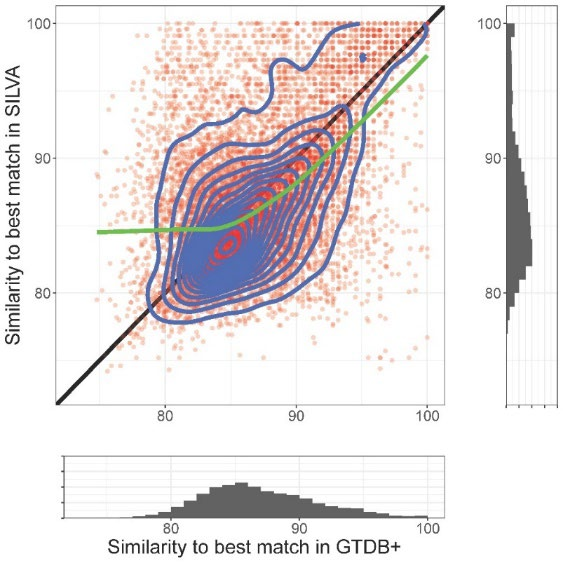
Our main aim in developing KSGP [citation] was to improve the taxonomic annotation of communities of Archaea by providing a database in which all sequences are assigned to an up to date taxonomic hierarchy based on coding genes and are provided with species level taxonomy for MAGs as well as cultivated strains. KSGP is also likely to be of use to those working with bacterial communities and an appendix of our paper briefly compares its performance on a bacterial data set with that of the recently released Greengenes2 database.
A
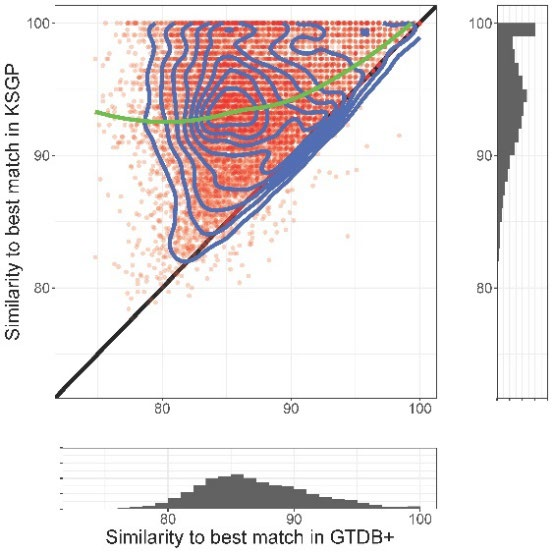
B
Edgar, R. C. (2018). Taxonomy annotation and guide tree errors in 16S rRNA databases. PeerJ, 6. doi:10.7717/peerj.5030
Karst, S. M., Dueholm, M. S., McIlroy, S. J., Kirkegaard, R. H., Nielsen, P. H., & Albertsen, M. (2018). Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nature Biotechnology, 36(2), 190-+. doi:10.1038/nbt.4045
Parks, D. H. (2023). Announcing-gtdb-r08-rs214. Retrieved from https://forum.gtdb.ecogenomic.org/t/announcing-gtdb-r08-rs214/456
Parks, D. H., Chuvochina, M., Chaumeil, P. A., Rinke, C., Mussig, A. J., & Hugenholtz, P. (2020). A complete domain-to-species taxonomy for Bacteria and Archaea. Nature Biotechnology, 38(9), 1079-+. doi:10.1038/s41587-020-0501-8
Parks, D. H., Chuvochina, M., Rinke, C., Mussig, A. J., Chaumeil, P.-A., & Hugenholtz, P. (2021). GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Research, 50(D1), D785-D794. doi:10.1093/nar/gkab776
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., & Hugenholtz, P. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nature Biotechnology, 36(10), 996-+. doi:10.1038/nbt.4229
Smith, R. H., Glendinning, L., Walker, A. W., & Watson, M. (2022). Investigating the impact of database choice on the accuracy of metagenomic read classification for the rumen microbiome. Animal Microbiome, 4(1). doi:10.1186/s42523-022-00207-7






